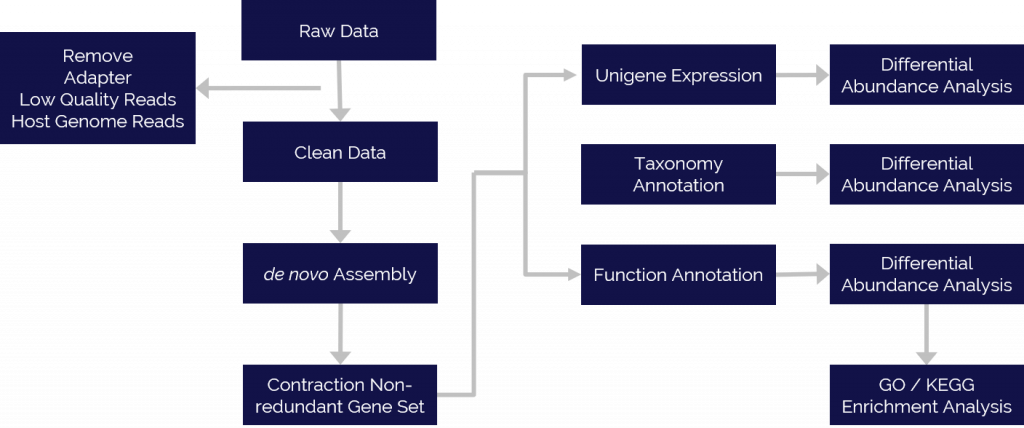
Because many natural microorganisms cannot be isolated and cultured or cloned, the vast majority of microbial biodiversity has been missed by traditional cultivation-based analysis methods. New next-gen DNA/RNA sequencing technologies enable us to directly sequence all genetic material that is recovered from all the members of a sampled community, including unculturable microorganisms that are otherwise difficult to analyze. This method provides, not only the species identification, classification and quantitation of microbes but also provides information for functional analysis of all the genes through gene ontology and metabolic pathway analysis.
Metatranscriptomic Sequencing
Technical Advantages
Comprehensive analysis – compared to 16S/18S/ITS sequencing, no selective amplification is used. All genetic material in the sample is sequenced. This method enables researchers to comprehensively sample all genes in all organisms present in a given complex sample.
No cloning – this technique does not require cloning the DNA before sequencing, removing one of the main biases and bottlenecks in environmental sampling.
All species – reporting includes all types of species diversity within your samples, including eukarya, archaea, bacteria and viruses.
Lower cost – as the price of Next-gen sequencing continues to fall, metatranscriptomics now allows microbial ecology to be investigated at a much greater scale and detail than before.