Whole-genome sequencing (WGS) is a comprehensive method for analyzing entire genomes.
Unlike focused approaches such as exome sequencing or targeted resequencing, which analyze a limited portion of the genome, whole-genome sequencing delivers a comprehensive view of the entire genome. It is ideal for discovery applications, such as identifying causative variants and novel genome assembly. Whole-genome sequencing can detect single nucleotide variants, insertions/deletions, copy number changes, and large structural variants.
Our Comprehensive Services Offer
Sample QC
Library preparation
High-throughput sequencing (Illumina next-gen sequencing technology)
Bioinformatics analysis
Customer data report
In-Depth Data Analysis Includes:
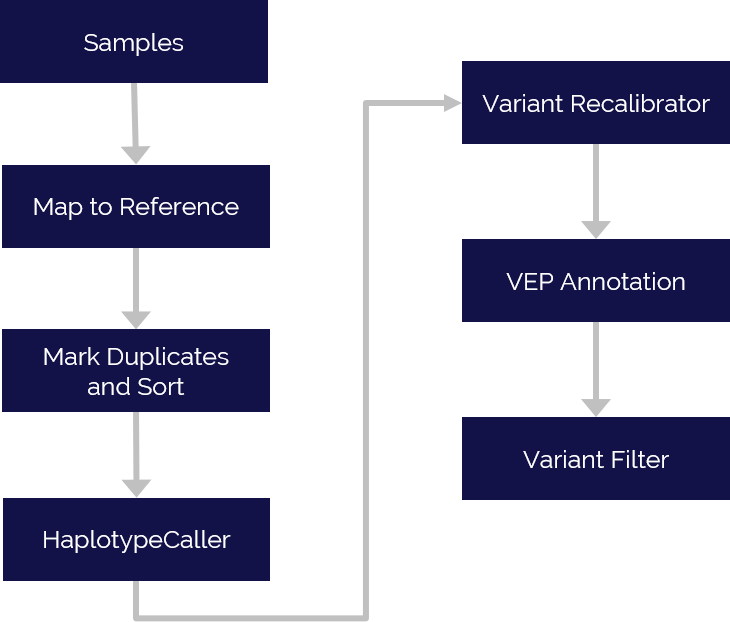
Assessment of sequencing and quality filtering of “junk” and low quality reads
Mapping reads to reference sequence database(s)
Performing on-target and coverage statistic analysis
Generating variant report including: single-nucleotide polymorphism (SNPs), single-nucleotide variations (SNVs), insertions and deletions (InDels), copy number variations (CNVs), and structural variations (SV)
Variant annotations with multiple databases including: ClinVar, dbNSFP, DisGeNET, OMIM, PheGenI, and VEP (annotations may be unavailable for non-human samples)
Differential mutations analysis between selected groups including: functional annotations and pathway analysis (KEGG)
Customer data report – includes a summary of methods and all statistic analysis
*Recommendations for human samples only. For non-human species, please consult with our technical support team.