Application Note
Researchers demonstrate that the intrinsically low raw accuracy of long-read sequencing technologies can be improved to a read recovery rate and crosstalk rate similar to current high-accuracy short-read sequencing technologies using non-random error-correcting barcodes synthesized in massive parallel on a microarray.
Error-correcting barcodes theory
NS-watermark barcodes, developed by researchers at the Centro Internacional Franco Argentino de Ciencias de la Información y de Sistemas, Argentina1, are inspired by watermark error-correcting codes, originally developed to deal with the issue of reliable transmission of information in the presence of noise in digital communications. The researchers have applied that theory here2 with the idea that barcodes of known sequence can be used not only to demultiplex reads, but also to correct sequencing errors and improve read recovery rate, if the barcode sequence contains the error-correcting codes.
Microarray synthesized oligo pools
The obvious advantages of massive parallel oligo synthesis over traditional column-based synthesis, such as scalability and lower cost, come with some challenges. Reduced scale of synthesis (attomoles per oligo) and the single pool of sequences format can be dealt with, but must be considered in the design of the oligo sequences. The researchers overcame these challenges by including primer annealing sites at either end of the oligonucleotides to allow their amplification via PCR.
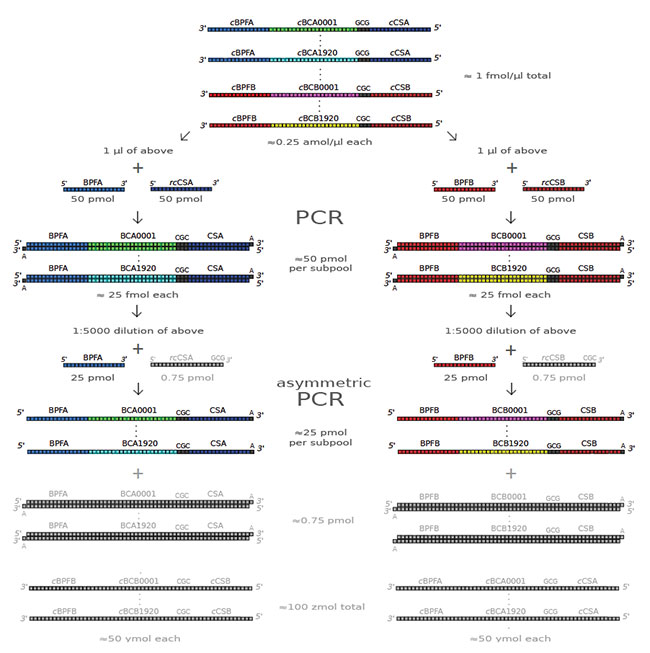
They designed 3840 distinct 36-bp variable barcode sequences and flanked them with two sets of fixed, 20-bp primer annealing regions which define two subpools A and B of 1920 oligos and used them to selectively amplify each subpool in two parallel PCR reactions. Double-stranded PCR product was converted to a single-stranded form, suitable for priming a downstream barcoding reaction using an asymmetric PCR (aPCR) reaction, where the forward primer is an ∼33:1 molar excess relative to the reverse primer, to preferentially amplify one of the strands.
Following ssDNA enrichment, these subpools are used as primers in two separate barcoding PCR reactions and then the barcoded products were pooled and subject to a single standard library preparation. The prepared library was sequenced on a MinION device and 1.6 million “pass” quality reads were obtained.
Two-round PCR separation and amplification of oligo subpools A and B
A first symmetric PCR selectively amplifies each subpool. A second asymmetric PCR preferentially amplifies the top strand. Except for the initial concentration, which was provided by the manufacturer, quantities shown are nominal and assume unitary reaction efficiencies.
Demultiplexing read recovery and crosstalk rate
The researchers used a two-level decoder (an LDPC iterative decoder algorithm in conjunction with a nucleotide-level Hidden Markov Model (HMM) predictor) for barcode demultiplexing of the reads. They analyzed the trade-off between read recovery rate (% of subreads which are not discarded by the decoder, low confidence, or as a negative control) and the crosstalk rate (% of recovered subreads which are assigned to an erroneous barcode) at increasingly stringent thresholds for the decoding confidence metric. They report a read recovery rate of 86.4% at a crosstalk rate of 0.17% (≈ 1 misassignment for every 584 demultiplexed reads).
Additionally, they report full representation of the microarray-synthesized oligonucleotides: all 3840 barcodes in subpools A and B were detected in at least 10 of the reads, while none of the 256 negative control barcodes was detected in more than 5 reads. Similarly, rich diversity was observed in terms of barcode combinations.
Thus, the researchers were able to demultiplex more than 86% of filtered reads tagged with 3840 barcodes with a crosstalk rate of 0.17%. This level of crosstalk is comparable to that exhibited by 96-plex single-end barcoding schemes in the Illumina platform despite the higher plexity and the combined error rate of: microarray-based synthesis, multiple rounds of PCR amplification using low-fidelity Taq polymerase and native, single-pass ONT sequencing.
Significant improvement
The number of distinct barcodes synthesized and demultiplexed here is one order of magnitude higher than the largest commercially-available barcoding kits for long-read sequencing (3840 vs. PacBio’s 384), while the diversity afforded by combinatorial asymmetric barcoding with this many distinct tags would be close to that offered by 12-nt unique molecular identifiers or UMIs (3840×3840≈14.7 M vs. 412≈16.7 M). Even higher numbers are easily within reach, as the NS-watermark barcode design framework is arbitrarily scalable (limited only by barcode length) and microarray-based synthesis of tens of thousands to millions of oligonucleotides per chip is already commercially available.
NS-watermark barcodes, particularly when combined with scalable, low-cost microarray-based synthesis, are a promising tool for long-read applications which require massive barcoding, high read recovery and low crosstalk.
OligoMix® has also been demonstrated as an effective method of high-throughput oligo synthesis for production of guide RNA libraries in CRISPR/Cas 9 high-throughput functional genomic screens
References
- Ezpeleta J, Krsticevic FJ, Bulacio P, Tapia E. (2017) Designing robust watermark barcodes for multiplex long-read sequencing. Bioinformatics 33(6):807-813. [article].
- Ezpeleta J, Garcia Labari I, Villanova GV, Bulacio P, Lavista-Llanos S, Posner V, Krsticevic F, Arranz S, Tapia E. (2022) Robust and scalable barcoding for massively parallel long-read sequencing. Sci Rep 12(1):7619. [article].