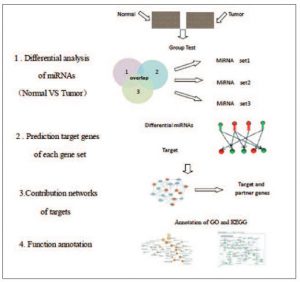
The aim of a recent study by researchers from Nanjing Medical University was to screen miRNAs related to different subtypes of breast cancer and their target genes, in order to identify new markers of tumor subtype. The miRNA expression profiles of breast cancer GSE38867 included 7 ductal carcinoma in situ breast (DCIS) cancer samples, 7 invasive breast cancer samples, 7 metastatic breast cancer samples, and 7 normal breast samples) which were downloaded from Gene Expression Omnibus (GEO) database. Profiling was performed using LC Sciences miRNA microarray service. Limma package in R software was applied to identify specific differentially expressed miRNAs of different subtypes of breast cancer and the microRNA.org database was the source used to predict target genes of the identified differentially expressed miRNAs. Researchers integrated the target genes and their interacted genes (predicted by STRING) into DAVID to perform the GO function and KEGG pathway analyses. Compared to their normal control, a total of 21, 47, and 107 differentially expressed miRNAs were screened in DCIS, invasive and metastatic breast cancer, respectively. Specific differentially expressed miRNAs of the three subtypes were identified, which included hsa-miR-99a and hsa-miR-151-3p for DCIS breast cancer, hsa-miR-145 and hsa-miR-210 for invasive breast cancer, and has-miR-205 and has-miR-361-5p metastatic breast cancer. Furthermore, 220, 43, 446, 307, 587 and 328 interaction pairs of the specific miRNA targets were predicted and it’s noted that multiple GO functions and KEGG pathways were enriched with the miRNA targets and their interacted genes. Through screening the most representative miRNAs of the three different subtypes of breast cancer, this team of researchers hope their data may act as putative markers in the diagnosis of different subtypes of breast cancer.
Diagram of prediction of putative breast cancer-related miRNAs based on the functional associations
Related Service
miRNA Microarray Service – LC Sciences provides a microRNA (miRNA) expression profiling service using microarrays based on our in-house developed µParaflo® technology platform. We have standard arrays for all mature miRNAs of all species available in the latest version of the miRBase database (Release 21, July 2014). Our service is comprehensive and includes sample labeling, array hybridization, image data processing and in-depth data analysis. Two-three weeks after receiving your total RNA samples, we’ll send you both the raw and fully analyzed data. [Learn more…]
Reference
E.-H. Sun, Q. Zhou, K.-S. Liu, W. Wer, C.-M. Wang, X.-F. Liu, C. Lu, D.-Y. Ma Screening miRNAs related to different subtypes of breast cancer with miRNAs microarray (2014) EurRevPharmacolSci 18: 2783-2788 [article]
