Researchers at Wenzhou Medical University set out to identify carcinogenic potential-related molecular mechanisms in cancer stem cells (CSCs) in lung cancer. Their study focused on adenocarcinomic human alveolar basal epithelial cells (A549 cells) and their expression of the CD133 antigen which is encoded by the PROM1 gene.
The researchers sorted CD133+ and CD133- subpopulations from A549 cells using magnetic-activated cell sorting and then compared abilities to form sphere and clone, proliferate, migrate, and invade, as well as drug sensitivity.
Next microRNA (miRNA) profiling was performed to identify differentially expressed miRNAs between CD133+ and CD133- subpopulations. LC Sciences carried out the microRNA microarray profiling and bioinformatics analysis to predict target genes for differentially expressed miRNAs and enrichment analysis. The researchers also explored mammalian target of rapamycin (mTOR) signaling pathways and CSC property-associated signaling pathways and visualized in regulatory network among competitive endogenous RNA (ceRNA), miRNA, and target gene.
They found that the CD133+ subpopulation showed greater oncogenic potential than CD133- subpopulation. In all, 14 differentially expressed miRNAs were obtained and enriched in 119 pathways, including five upregulated and nine downregulated. For mTOR signaling pathway, eight differential miRNAs and 39 target genes were involved, as well as some ceRNAs. For CSC property-related signaling pathways, six miRNAs were dramatically enriched in Hedgehog, Notch, and Wnt signaling pathways via regulating 108 target genes.
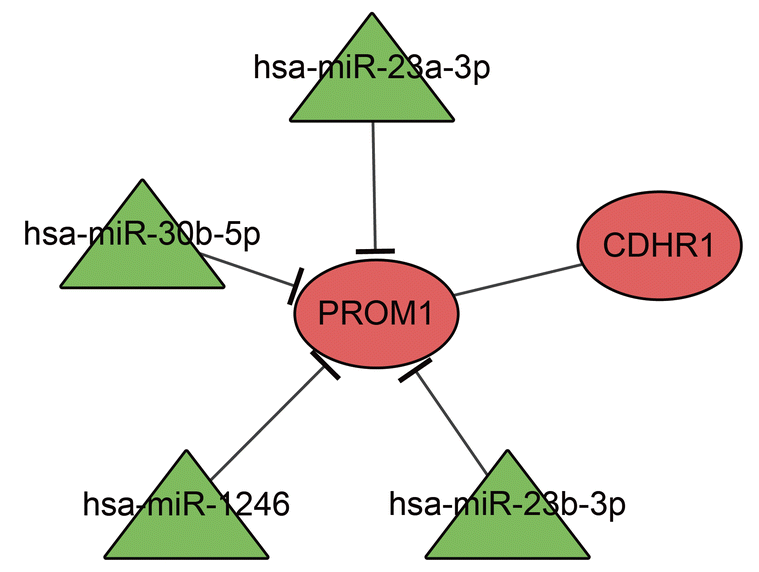
Network of differentially expressed miRNA and CD133 (PROM1). Red circle represents target gene, and green triangle represents differentially expressed miRNA. miRNA, microRNA.
The mTOR and CSC property-associated signaling pathways may be important oncogenic molecular mechanisms in CD133+ A549 cells.
Related Service
miRNA Microarray Service – LC Sciences provides a microRNA (miRNA) expression profiling service using microarrays based on our in-house developed µParaflo® technology platform. We have standard arrays for all mature miRNAs of all species available in the latest version of the miRBase database (Release 21, July 2014). Our service is comprehensive and includes sample labeling, array hybridization, image data processing and in-depth data analysis. Two-three weeks after receiving your total RNA samples, we’ll send you both the raw and fully analyzed data. [Learn more…]
Reference
Chen QY, Jiao DM, Zhu Y, Hu H, Wang J, Tang X, Chen J, Yan L. (2015) Identification of carcinogenic potential-associated molecular mechanisms in CD133+ A549 cells based on microRNA profiles. Tumour Biol 37(1):521-30. [abstract]