Alzheimer’s disease (AD) is a uniquely human, age-related central nervous system (CNS) disorder for which there is no adequate experimental model. While well over 100 transgenic murine models of AD (TgAD) have been developed that recapitulate many of the neuropathological features of AD, key pathological features of AD such as progressive neuronal atrophy, neuron cell loss, and neurofibrillary tangle (NFT) formation have not been observed in any TgAD model to date. To more completely analyze and understand the neuropathology, altered neuro-inflammatory and innate-immune signaling pathways, and the complex molecular-genetics and epigenetics of AD, it is therefore necessary to rigorously examine short post-mortem interval (PMI) human brain tissues to gain a deeper and more thorough insight into the neuropathological mechanisms that characterize the AD process. A recent perspective-methods paper published by researchers from Louisiana State University highlights some important recent findings on the utilization of short PMI tissues in sporadic (idiopathic; of unknown origin) AD research with focus on the extraction and quantification of RNA, and in particular microRNA (miRNA) and messenger RNA (mRNA) and analytical strategies, drawing on the authors’ combined 125 years of laboratory experience into this investigative research area. It’s their hope that new investigators in the field of “gene expression analysis in neurological disease” will benefit from the observations presented in this paper and incorporate these recent findings and observations into their future experimental planning and design.
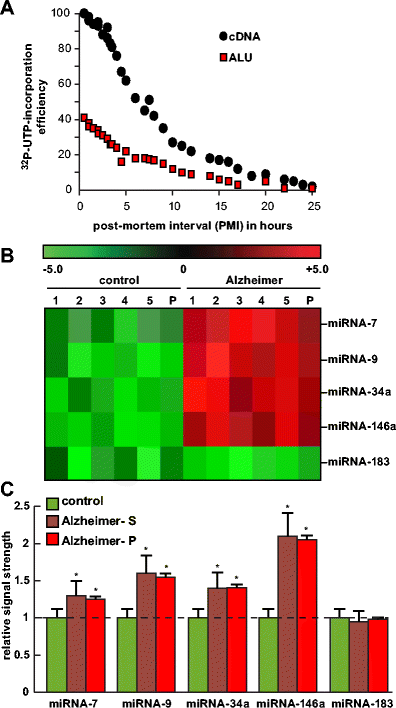
a Analysis of RNA integrity in short post-mortem interval (PMI) human brain tissues (superior temporal lobe neocortex)—relative efficiency of run-on gene transcription as a function of PMI (measured in hours) using total cDNA or ALU DNA probes. Briefly, as an index of human brain total RNA quality, run-on transcription was performed in human neocortical nuclei isolated from 0.5- to 25-h PMI human brains using [α-32P]-UTP label incorporation into newly synthesized RNA message, as assessed by probing with either total cDNA or ALU repeat DNA probes on northern dot blot membranes . ALU elements represent one of the most highly repetitive ∼300 base pair repeats in the human genome, and the presence of transcribed ALU sequences in RNA can be used as an index of transcriptional output of newly synthesized RNA . The efficiency of run-on gene transcription appears to decrease markedly after a 3- to 4-h PMI. Interestingly, a comparison between either total cDNA or ALU DNA probes vs 0.5- to 25-h PMI shows parallel signal decay kinetics utilizing the ALU probe, yielding approximately 30–40 % of the total cDNA signal at any given PMI. Analysis of short PMI brain using run-on gene transcription may be useful for genetic activity studies in characterizing the transcriptional competency of brain cells during normal human aging and in neurodegenerative diseases such as AD; extensive experimental details have been previously described . b Individual and pooled analysis of samples in the human brain (superior temporal lobe neocortex)–miRNA analysis; individual age-matched control (control; N = 5) or Alzheimer’s disease (Alzheimer; N = 5) samples or pools of these samples (P) were analyzed using miRNA arrays (LC Sciences, Houston, TX); specific data for human miRNA-7, miRNA-9, miRNA-34a, miRNA-146a, and an unchanging (control) miRNA-183 were selected for analysis. c Compared to control, the mean signal strength of five single-AD samples (Alzheimer–S) gave comparable signal yields to a single pooled sample containing these same five single samples (Alzheimer-P); pooling of samples has the significant advantage of being more cost-effective, and individual agonal processes are more effectively controlled to emphasize AD-relevant trends (see text); pooling of samples may be further useful in the analysis of gene expression studies involving miRNA and/or mRNA in human population studies; as the colored bar graph indicates miRNA-7, miRNA-9, miRNA-34a, and miRNA-146a are significantly upregulated in AD over age-matched controls; miRNA-34a has been recently implicated in the downregulation of TREM2 expression in AD brain and an inability to effectively phagocytose Aβ peptides; data are the mean ± one standard deviation of that mean; *p < 0.05 (ANOVA)
Related Service
miRNA Microarray Service – LC Sciences provides a microRNA (miRNA) expression profiling service using microarrays based on our in-house developed µParaflo® technology platform. We have standard arrays for all mature miRNAs of all species available in the latest version of the miRBase database (Release 21, July 2014). Our service is comprehensive and includes sample labeling, array hybridization, image data processing and in-depth data analysis. Two-three weeks after receiving your total RNA samples, we’ll send you both the raw and fully analyzed data. [Learn more…]
Reference
Clement C, Hill JM, Dua P, Culicchia F, Lukiw WJ. (2015) Analysis of RNA from Alzheimer’s Disease Post-mortem Brain Tissues Mol Neuro 53(2):1322-8. [article]
