Variation in gene expression is an important mechanism underlying susceptibility to complex disease and traits. Single nucleotide polymorphisms (SNPs) account for a substantial portion of the total detected genetic variation in gene expression but how exactly variants acting in trans modulate gene expression and disease susceptibility remains largely unknown. The BDNF Val66Met SNP has been associated with a number of psychiatric disorders such as depression, anxiety disorders, schizophrenia and related traits.
Using LC Sciences’ global microRNA expression profiling in hippocampus of humanized BDNF Val66Met knock-in mice Columbia University researchers showed that this variant results in dysregulation of at least one microRNA, which in turn affects downstream target genes. Specifically, we show that reduced levels of miR-146b (mir146b), lead to increased Per1 and Npas4 mRNA levels and increased Irak1 protein levels in vitro and are associated with similar changes in the hippocampus of hBDNFMet/Met mice. Our findings highlight trans effects of common variants on microRNA-mediated gene expression as an integral part of the genetic architecture of complex disorders and traits.
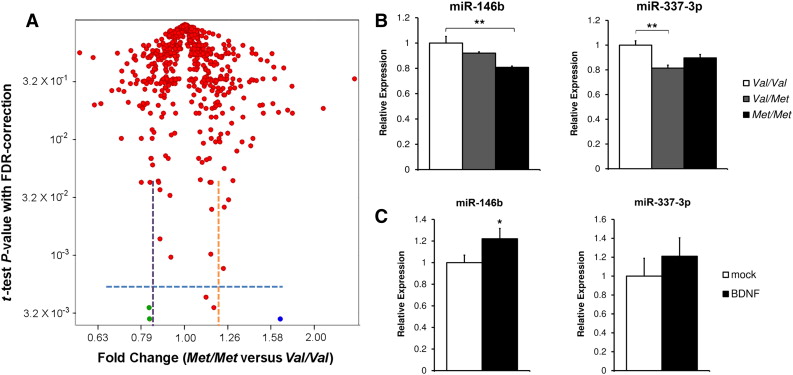
miR-146b expression levels are decreased in the hippocampus of hBDNFMet/Met mice. (A) miRNA expression profile in hippocampus of hBDNFMet/Met animals versus hBDNFVal/Val animals (n = 5 each genotype). Volcano plot showing the FDR-corrected P-value and corresponding relative expression of each miRNA. Green dots, miR-146b and miR-337-3p; blue dot, miR-700. (B) Expression levels of miR-146b and miR-337-3p in hippocampus of hBDNFVal/Val, hBDNFVal/Met and hBDNFMet/Met mice (n = 6 each genotype), as measured by qRT-PCR. Expression levels in hBDNFVal/Met and hBDNFMet/Met mice were normalized to hBDNFVal/Val animals. (C) Expression levels of miR-146b and miR-337-3p in acute hippocampal slices (n = 9, from 3 animals, each treatment) treated with BDNF for 2 h, as measured by qRT-PCR. Expression levels following BDNF treatments were normalized to mock treatment control. Results are expressed as mean ± SEM. *P < 0.05, **P < 0.01, ANOVA with post-hoc Bonferroni’s test (B); Student’s t-test (C).
Related Service
miRNA Microarray Service – LC Sciences provides a microRNA (miRNA) expression profiling service using microarrays based on our in-house developed µParaflo® technology platform. We have standard arrays for all mature miRNAs of all species available in the latest version of the miRBase database (Release 21, July 2014). Our service is comprehensive and includes sample labeling, array hybridization, image data processing and in-depth data analysis. Two-three weeks after receiving your total RNA samples, we’ll send you both the raw and fully analyzed data. [Learn more…]
Reference
Hsu P, Xu B, Mukai J, Karayiorgou M, Gogos JA. (2015) The BDNF Val66Met variant affects gene expression through miR-146b. Neurobiology of Disease 77(1), 228-237. [article]