Diabetic polyneuropathy (DPN) is a common but irreversible neurodegenerative complication of diabetes mellitus. In a recent paper published by a team of scientists led by researchers from the University of Alberta, investigators show that features of sensory neuron damage in mice with chronic DPN may have altered epigenetic micro RNA (miRNA) transcriptional control. In their work, they profiled sensory neuron messenger RNA and miRNA profiles in mice with type I diabetes mellitus and findings of DPN. Diabetic sensory dorsal root ganglia neurons showed a pattern of altered messenger RNA profiles associated with upregulated cytoplasmic sites of miRNA-mediated messenger RNA processing (GW/P bodies). Using LC Sciences’ miRNA microarray service, researchers identified significant changes in expression among mice with diabetes, the most prominent of which were a 39% downregulation of mmu-let-7i and a 255% increase in mmu-miR-341; both were identified in sensory neurons.
To counteract these alterations, investigators replenished let-7i miRNA by intranasal administration; in a separate experiment, they added an anti-miR that antagonized elevated mmu-341 after 5 months of diabetes. Both approaches were shown to independently improve electrophysiologic, structural, and behavioral abnormalities without altering hyperglycemia; control sequences did not have those effects. Researchers found that dissociated adult sensory neurons exposed to an exogenous mmu-let-7i mimic displayed enhanced growth and branching, indicating a trophic action. These findings identify roles for epigenetic miRNA alterations and enhanced GW/P expression in diabetic dorsal root ganglia that contribute to the complex DPN phenotype.
Heat map results comparing DRG miRNA microarrays of 5-month-old mice with diabetes and mice without diabetes at the p < 0.05(a) and p < 0.01 significance levels(b).
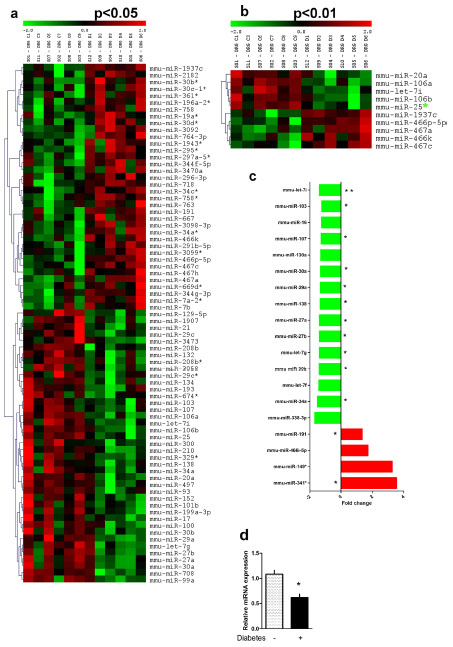
(c) Selected higher abundance miRNAs are shown indicating fold change and significance level at p < 0.10. * p < 0.05 and ** p < 0.01, Student t-test; n = 6 control mice; n = 6 mice with diabetes (for each mouse DRG, n = 1). (d) Independent miRNA qRT-PCR analysis of relative mmu-let-7i levels confirms its reduction in diabetic DRG. * p = 0.001, Student t-test; n = 6 nondiabetic arrays; n = 5 diabetic arrays.
Related Service
miRNA Microarray Service – LC Sciences provides a microRNA (miRNA) expression profiling service using microarrays based on our in-house developed µParaflo® technology platform. We have standard arrays for all mature miRNAs of all species available in the latest version of the miRBase database (Release 21, July 2014). Our service is comprehensive and includes sample labeling, array hybridization, image data processing and in-depth data analysis. Two-three weeks after receiving your total RNA samples, we’ll send you both the raw and fully analyzed data. [Learn more…]
Reference
Cheng C, Kobayashi M, Martinez JA, Ng H, Moser JJ, Wang X, Singh V. Fritzler MJ, Zochodne DW. (2015) Evidence for Epigenetic Regulation of Gene Expression and Function in Chronic Experimental Diabetic Neuropathy J Neuropathol Exp Neurol 74(8):804-17 [article]
