Background
RDX is a well-known pollutant to induce neurotoxicity. MicroRNAs (miRNA) and messenger RNA (mRNA) profiles are useful tools for toxicogenomics studies. It is worthy to integrate MiRNA and mRNA expression data to understand RDX-induced neurotoxicity.
Results
Researchers from Rush University Medical Center treated rats with or without RDX for 48 h. Both miRNA and mRNA profiles were conducted using brain tissues. The miRNA microarray analysis was performed by LC Sciences. Nine miRNAs were significantly regulated by RDX. Of these, 6 and 3 miRNAs were up- and down-regulated respectively. The putative target genes of RDX-regulated miRNAs were highly nervous system function genes and pathways enriched. Fifteen differentially genes altered by RDX from mRNA profiles were the putative targets of regulated miRNAs. The induction of miR-71, miR-27ab, miR-98, and miR-135a expression by RDX, could reduce the expression of the genes POLE4, C5ORF13, SULF1 and ROCK2, and eventually induce neurotoxicity. Over-expression of miR-27ab, or reduction of the expression of unknown miRNAs by RDX, could up-regulate HMGCR expression and contribute to neurotoxicity. RDX regulated immune and inflammation response miRNAs and genes could contribute to RDX- induced neurotoxicity and other toxicities as well as animal defending reaction response to RDX exposure.
Conclusions
These results demonstrate that integrating miRNA and mRNA profiles is valuable to indentify novel biomarkers and molecular mechanisms for RDX-induced neurological disorder and neurotoxicity.
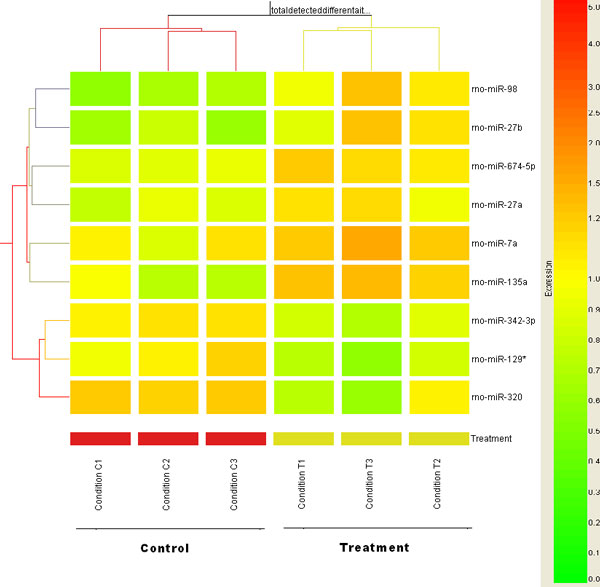
Hierarchical clustering of differentially expressed miRNAs induced by RDX
Nine differentially expressed miRNAs (horizontal axis) were used for a Two-Way hierarchical clustering across all the control and RDX treated samples (vertical axis). A Pearson correlation algorithm was applied to calculate the distances between transcripts or between conditions. The relative level of gene expression is indicated by the color scale at the right side.
Related Service
miRNA Microarray Service – LC Sciences provides a microRNA (miRNA) expression profiling service using microarrays based on our in-house developed µParaflo® technology platform. We have standard arrays for all mature miRNAs of all species available in the latest version of the miRBase database (Release 21, July 2014). Our service is comprehensive and includes sample labeling, array hybridization, image data processing and in-depth data analysis. Two-three weeks after receiving your total RNA samples, we’ll send you both the raw and fully analyzed data. [Learn more…]
Reference
Y Deng, J Ai, X Guan, Z Wang, B Yan, D Zhang, C Liu, M Wilbanks, B Escalon, S Meyers, M Yang, E Perkins (2014) MicroRNA and messenger RNA profiling reveals new biomarkers and mechanisms for RDX induced neurotoxicity BMC Genomics 15(Suppl 11):S1 doi:10.1186/1471-2164-15-S11-S1 [article]