Owing to the dynamic nature of the transcriptome, gene expression profiling is a promising tool for discovery of disease-related genes and biological pathways. In the present study, researchers at the University of Glasgow examined gene expression in whole blood of 12 patients with CAD (coronary artery disease) and 12 healthy control subjects. Furthermore, ten patients with CAD underwent whole-blood gene expression analysis before and after the completion of a cardiac rehabilitation programme following surgical coronary revascularization. mRNA and miRNA (microRNA) were isolated for expression profiling. Gene expression analysis identified 365 differentially expressed genes in patients with CAD compared with healthy controls (175 up- and 190 down-regulated in CAD), and 645 in CAD rehabilitation patients (196 up- and 449 down-regulated post-rehabilitation). Biological pathway analysis identified a number of canonical pathways, including oxidative phosphorylation and mitochondrial function, as being significantly and consistently modulated across the groups. Analysis of miRNA expression by LC Sciences revealed a number of differentially expressed miRNAs, including hsa-miR-140-3p (control compared with CAD, P=0.017), hsa-miR-182 (control compared with CAD, P=0.093), hsa-miR-92a and hsa-miR-92b (post- compared with pre-exercise, P<0.01). Global analysis of predicted miRNA targets found significantly reduced expression of genes with target regions compared with those without: hsa-miR-140-3p (P=0.002), hsa-miR-182 (P=0.001), hsa-miR-92a and hsa-miR-92b (P=2.2×10-16).
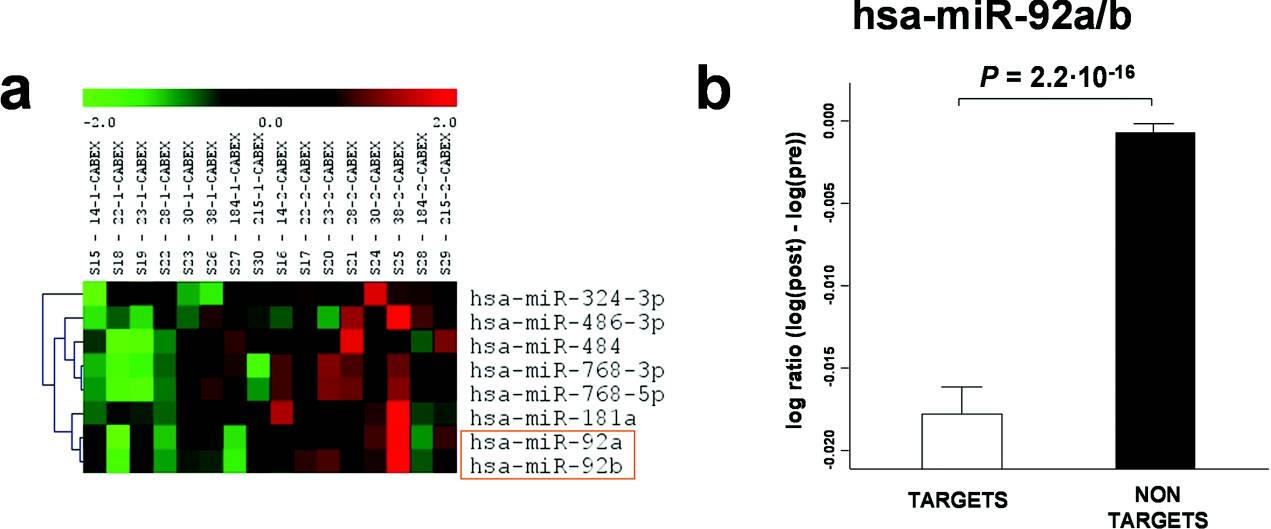
miRNA expression in patients with CAD before and after completion of the cardiac rehabilitation programme
(a) Heatmap analysis of miRNA expression in patients with CAD before and after completion of the cardiac rehabilitation programme (P<0.10; signal intensity >500 intensity units). The analysis shows relative fluorescence from green (2-fold reduction) to red (2-fold increase) of each miRNA per individual (with anonymous identifier at the top of each column). (b) Comparison of log ratios of hsa-miR-92a/b identified a significant reduction in gene expression levels in predicted target genes compared with non-targets.
In conclusion, using whole blood as a ‘surrogate tissue’ in patients with CAD, the researchers have identified differentially expressed miRNAs, differentially regulated genes and modulated pathways which warrant further investigation in the setting of cardiovascular function. This approach may represent a novel non-invasive strategy to unravel potentially modifiable pathways and possible therapeutic targets in cardiovascular disease.
Related Service
miRNA Microarray Service – LC Sciences provides a microRNA (miRNA) expression profiling service using microarrays based on our in-house developed µParaflo® technology platform. We have standard arrays for all mature miRNAs of all species available in the latest version of the miRBase database (Release 21, July 2014). Our service is comprehensive and includes sample labeling, array hybridization, image data processing and in-depth data analysis. Two-three weeks after receiving your total RNA samples, we’ll send you both the raw and fully analyzed data. [Learn more…]
Reference
Taurino C, Miller WH, McBride MW, McClure JD, Khanin R, Moreno MU, Dymott JA, Delles C, Dominiczak AF. (2010) Gene expression profiling in whole blood of patients with coronary artery disease. Clin Sci 119(8), 335-43. [article]