Dissecting the gene expression programs which control the early stage cardiovascular development is essential for understanding the molecular mechanisms of human heart development and heart disease. In a recent paper published by researchers from University of Pittsburgh School of Medicine, investigators emplyed LC Sciences’ RNA-sequencing service to profile the transcriptome of highly purified human Embryonic Stem Cells (hESCs), hESC-derived Multipotential Cardiovascular Progenitors (MCPs) and MCP-specified three cardiovascular lineages. A novel algorithm, named as Gene Expression Pattern Analyzer (GEPA), was developed to obtain a refined lineage-specificity map of all sequenced genes, which reveals dynamic changes of transcriptional factor networks underlying early human cardiovascular development. Moreover, their GEPA predictions captured ~90% of top-ranked regulatory cardiac genes that were previously predicted based on chromatin signature changes in hESCs, and further defined their cardiovascular lineage-specificities, indicating that our multi-fate comparison analysis could predict novel regulatory genes. Furthermore, GEPA analysis revealed the MCP-specific expressions of genes in ephrin signaling pathway, positive role of which in cardiomyocyte differentiation was further validated experimentally. By using RNA-seq plus GEPA workflow, researchers also identified stage-specific RNA splicing switch and lineage-enriched long non-coding RNAs during human cardiovascular differentiation. Overall, this study utilized multi-cell-fate transcriptomic comparison analysis to establish a lineage-specific gene expression map for predicting and validating novel regulatory mechanisms underlying early human cardiovascular development.
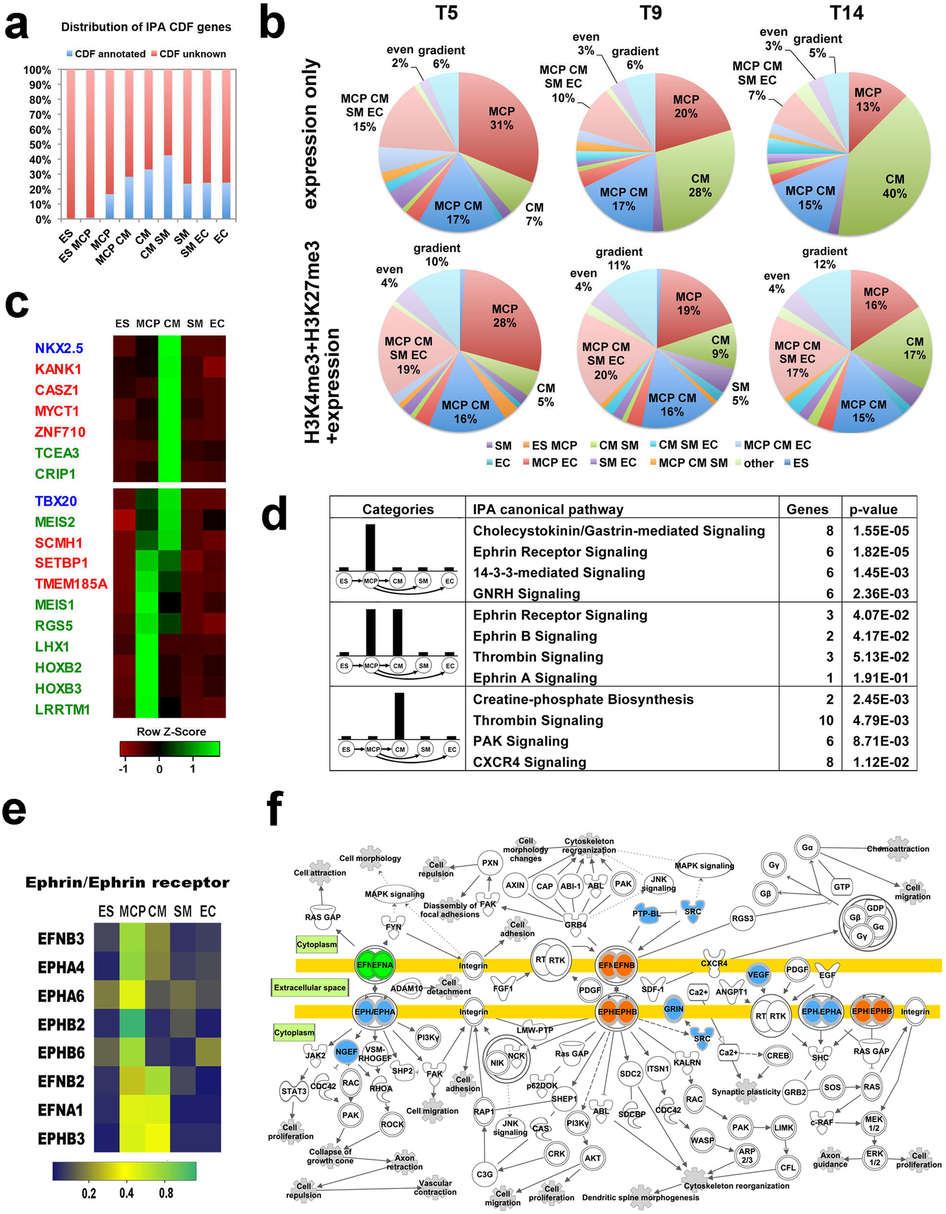
(a) Percent of the genes with or without annotation in “cardiovascular development and function” from Ingenuity knowledge database in the LEG groups identified by GEPA. (b) Our GEPA analysis of top 100 novel cardiac regulatory genes previously predicted by Paige et al.when considering “expression only” or “H3K4me3+H3K27me3+expression “ at T5, T9 and T14 of CM differentiation, respectively. (c) Examples of predicted functional genes by GEPA. Dynamic expression of these genes is shown in heatmap. (d) Predicted novel regulatory pathways in cardiovascular differentiation using GEPA and Inginuity IPA pathway enrichment analysis. (e) A heat-map showing the lineage-specific expression pattern of ephrin and ephrin receptor genes during cardiovascular differentiation. (f) Illustrative Ephrin/Ephrin signaling pathway imposed on a pathway map based on Ingenuity IPA showing localizations of the LEGs.
LEGs identified by GEPA predict novel regulatory genes and pathways in human cardiovascular differentiation
Related Service
RNA Sequencing Service – LC Sciences provides a one-stop solution (i.e. from sample to data) for RNA-Seq using the latest in rna sequencing technology. The RNA-Seq services provided by LC Sciences are comprehensive and provide the most complete picture of RNA content in your samples. We can help you set up and conduct a high-quality, well-controlled RNA-Seq experiment based on the latest deep-sequencing technologies.[Learn more…]
Reference
Y. Li, B. Lin, L. Yang (2015) Comparative Transcriptomic Analysis of Multiple Cardiovascular Fates from Embryonic Stem Cells Predicts Novel Regulators in Human Cardiogenesis Scientific Reports 5:9758 doi: 10.1038/srep09758 [article]