Glucose and oxygen deprivation during ischemia is known to affect the homeostasis of the endoplasmic reticulum (ER) in ways predicted to activate the unfolded protein response (UPR). Activation of UPR signalling due to ER stress is associated with the development of myocardial infarction (MI). MicroRNAs (miRNAs) are key regulators of cardiovascular development and deregulation of miRNA expression is involved in the onset of many cardiovascular diseases. However, little is known about the mechanisms regulating the miRNA expression in the cardiovascular system during disease development and progression. Therefore, a team led by researchers from the National University of Ireland Galway performed genome-wide miRNA expression profiling in rat cardiomyoblasts to identify the miRNAs deregulated during UPR, a crucial component of ischemia. Through the use of LC Sciences miRNA microarray service, researchers found that expression of 86 microRNAs changed significantly during conditions of UPR in H9c2 cardiomyoblasts. The team found that miRNAs with known function in cardiomyoblasts biology (miR-206, miR-24, miR-125b, miR-133b) were significantly deregulated during the conditions of UPR in H9c2 cells. The expression of miR-7a was upregulated by UPR and simulated in vitro ischemia in cardiomyoblasts. Further, ectopic expression of miR-7a provides resistance against UPR-mediated apoptosis in cardiomyoblasts. The ample overlap of miRNA expression signature between this analysis and different models of cardiac dysfunction further confirms the role of UPR in cardiovascular diseases. The study demonstrates the role of UPR in deregulating the expression of miRNAs in MI. The results provide novel insights about the molecular mechanisms of deregulated miRNA expression during the heart disease pathogenesis.
miRNA expressions during unfolded protein response in H9c2 cardiomyocytes
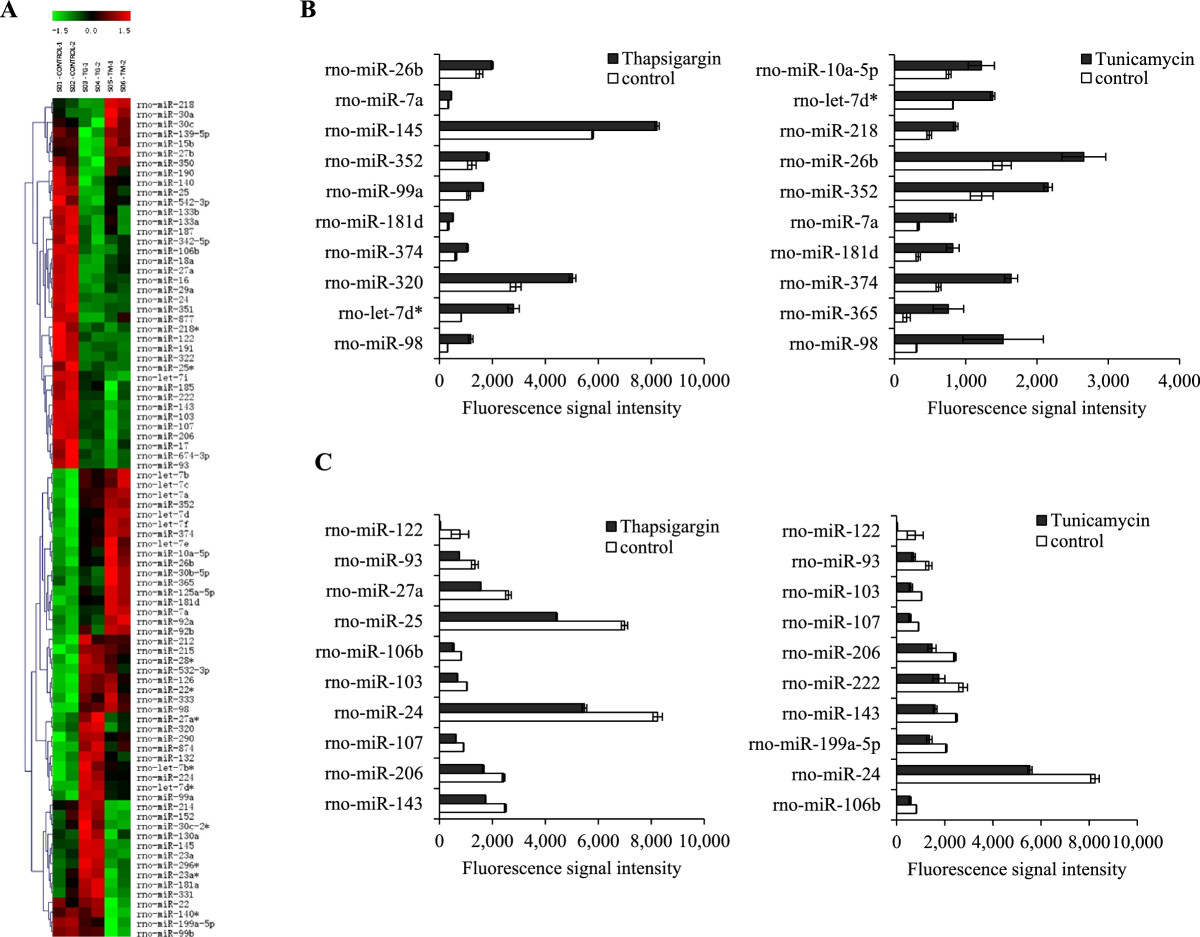
(A) Heat map summarizing the expression of miRNAs. Each column represents 1 of the samples, and each row represents 1 of 86 differentially expressed miRNAs at P <0.05. Samples were grouped by treatments, and miRNAs arranged by unsupervised hierarchical clustering. Red and green indicate up and downregulation, respectively, relative to the overall mean for each miRNA. Control-1 and control-2, untreated H9c2 cells; Tg-1 and Tg-2, thapsigargin treated H9c2 cells; Tm-1 and Tm-2, tunicamycin treated H9c2 cells. (B) Expression pattern of top 10 miRNAs (in terms of fluorescence intensity) upregulated upon treatment with thapsigargin and tunicamycin in H9c2 cells. (C) Expression pattern of top 10 miRNAs (in terms of fluorescence intensity) downregulated upon treatment with thapsigargin and tunicamycin in H9c2 cells.
Related Service
miRNA Microarray Service – LC Sciences provides a microRNA (miRNA) expression profiling service using microarrays based on our in-house developed µParaflo® technology platform. We have standard arrays for all mature miRNAs of all species available in the latest version of the miRBase database (Release 21, July 2014). Our service is comprehensive and includes sample labeling, array hybridization, image data processing and in-depth data analysis. Two-three weeks after receiving your total RNA samples, we’ll send you both the raw and fully analyzed data. [Learn more…]
Reference
Danielle Read, Ananya Gupta, Yury Ladilov, Afshin Samali, Sanjeev Gupta miRNA signature of unfolded protein response in H9c2 rat cardiomyoblasts (2014) Cell & Bioscience 2014, 4:56 doi:10.1186/2045-3701-4-56 [article]