Cardiac sarco(endo)plasmic reticulum calcium ATPase-2 (SERCA2) plays one of the central roles in myocardial contractility. Both, SERCA2 mRNA and protein are reduced in myocardial infarction (MI), but the correlation has not been always observed. MicroRNAs (miRNAs) act by targeting 3′-UTR mRNA, causing translational repression in physiological and pathological conditions, including cardiovascular diseases. Researchers at the University of Ljubljana aimed to identify miRNAs that could influence SERCA2 expression in human MI.
The protein SERCA2 was decreased and 43 miRNAs were deregulated in infarcted myocardium compared to corresponding remote myocardium, analyzed by western blot and microRNA microarrays, respectively. All the samples were stored as FFPE tissue and in RNAlater. miRNAs binding prediction to SERCA2 including four prediction algorithms (TargetScan, PicTar, miRanda and mirTarget2) identified 213 putative miRNAs. TAM and miRNApath annotation of deregulated miRNAs identified 18 functional and 21 diseased states related to heart diseases, and association of the half of the deregulated miRNAs to SERCA2. Free-energy of binding and flanking regions (RNA22, RNAfold) was calculated for 10 up-regulated miRNAs from microarray analysis (miR-122, miR-320a/b/c/d, miR-574-3p/-5p, miR-199a, miR-140, and miR-483), and nine miRNAs deregulated from microarray analysis were used for validation with qPCR (miR-21, miR-122, miR-126, miR-1, miR-133, miR-125a/b, and miR-98). Based on qPCR results, the comparison between FFPE and RNAlater stored tissue samples, between Sybr Green and TaqMan approaches, as well as between different reference genes were also performed.
Combing all the results, the researchers identified certain miRNAs as potential regulators of SERCA2; however, further functional studies are needed for verification. Using qPCR, we confirmed deregulation of nine miRNAs in human MI, and show that qPCR normalization strategy is important for the outcome of miRNA expression analysis in human MI.
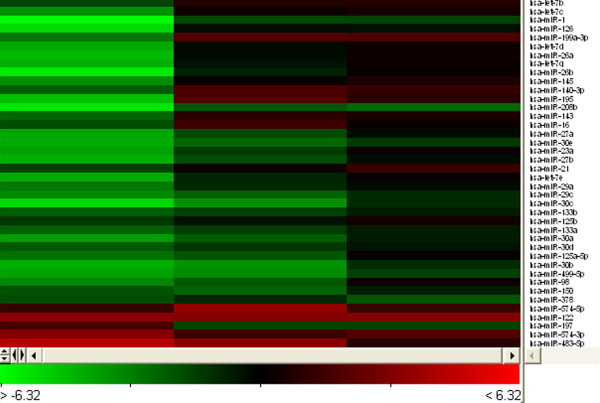
Dual colour experiments and swap-dye were performed, using Cy3 and Cy5 to compare miRNAs expression in infarcted tissue and corresponding remote myocardium. Statistical analysis and hierarchical clustering was performed using Acuity 4.0 analytical software. The results are displayed in a heat map. The legend on the right indicates the miRNA shown in the corresponding row, all of which are human specific. The bar code on the bottom shows the colour scale of the log2 value. Each column represents data from one microarray. The heat map was constructed of 43 miRNAs dysregulated in all of the microarrays performed.
Related Service
miRNA Microarray Service – LC Sciences provides a microRNA (miRNA) expression profiling service using microarrays based on our in-house developed µParaflo® technology platform. We have standard arrays for all mature miRNAs of all species available in the latest version of the miRBase database (Release 21, July 2014). Our service is comprehensive and includes sample labeling, array hybridization, image data processing and in-depth data analysis. Two-three weeks after receiving your total RNA samples, we’ll send you both the raw and fully analyzed data. [Learn more…]
Reference
Boštjančič E, Zidar N, Glavač D. (2012) MicroRNAs and cardiac sarcoplasmic reticulum calcium ATPase-2 in human myocardial infarction: expression and bioinformatic analysis. BMC Genomics 13,552. [article]