Annual teosinte, the ancestor of cultivated maize (Zea mays ssp. mays), is a valuable germplasm for enhancing the genetic diversity and adaptability of maize to various environment stimuli. However, comprehensive transcriptomic and genomic resources are unavailable in public databases, which significantly impede the identification and utilization of favourable genes or alleles in teosinte.
In this study, researchers at Huazhong Agricultural University globally sequenced the transcriptomes of six teosinte accessions by Illumina paired-end sequencing. A high-quality teosinte transcriptome was de novo assembled with an average length of 770 bp and 63.99% of annotated unigenes. They found approximately 75% of the genes were highly conserved between maize and teosinte. Moreover, the researchers also found 1516 unigenes were specifically expressed in teosinte, of which 84 unigenes were supported by gene models of four plant species, and 571 unigenes were located in the intergenic regions of maize genome, showing evidence-based expressed presence/absence variations (ePAVs). Furthermore, they also identified 99 unigenes with strong selection signals and 57 unigenes with >1 Ka/Ks ratios, suggesting that these genes might be under strong selection during maize domestication and improvement. Additionally, 11 286 teosinte unigene-derived primer pairs were developed for amplifying simple sequence repeat (SSR) loci.
Gene ontology (GO) analysis of the teosinte transcriptome
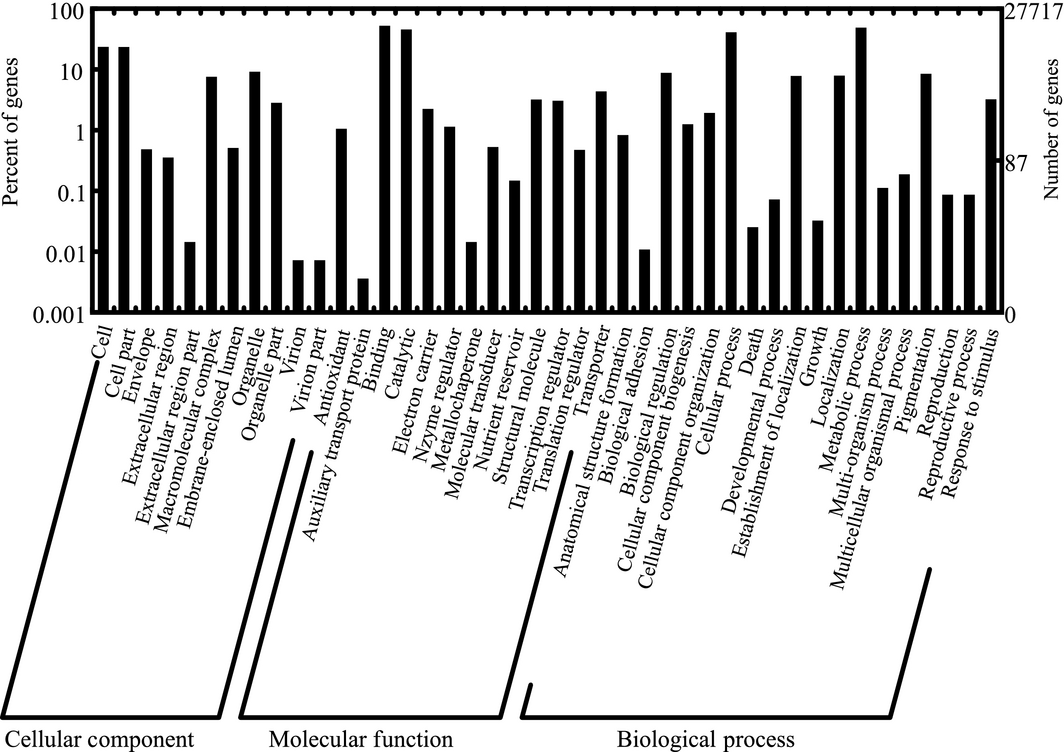
A total of 27 717 unigenes are assigned to three ontologies with 42 functional terms. The left y-axis represents the percentage of unigenes of a specific subcategory in each main category. The right y-axis indicates the number of unigenes in a subcategory.
This study provides a comprehensive transcriptome of teosinte, a subset of genes that are highly conserved or diverged during maize domestication and improvement, and a massive set of available SSR primer pairs. These results facilitate the investigation of the comparative genomics and molecular domestication of teosinte and the utilization of teosinte germplasm for maize improvement.
Related Service
Poly(A) RNA Sequencing Service – RNA-Seq technology is providing fresh biological insight into the transcriptome. At its most powerful, it can be used to determine the structure of genes, their splicing patterns and other post transcriptional modifications, to detect rare and and novel transcripts, and to quantify the changing expression levels of each transcript. [Learn more…]
References
Huang J, Gao Y, Jia H, Zhang Z. (2016) Characterization of the teosinte transcriptome reveals adaptive sequence divergence during maize domestication. Molecular Ecology Resources 16(6), 1465-1477 [abstract]
