A team led researchers at the Minzu University of China highlight the utility of high-throughput transcriptome sequencing as a fast and cost-effective approach for marker development and gene discovery in non-model species.
Ammopiptanthus mongolicus is an ecologically important plant species in Mid-Asia desert and exhibits substantial tolerance to drought condition. Insufficient transcriptomic and genomic data in public databases has limited our understanding of the molecular mechanism underlying the stress tolerance of A. mongolicus. They used 454 pyrosequencing to identify assembled sequences representing approximately 9,771 proteins in PlantGDB. The 29,056 unique sequences in this 454EST collection represent a major transcriptomic level resource for A. mongolicus, and will be useful for further functional genomics study in Ammopiptanthus genus. The thousands of SSR markers predicted in our 454 ESTs should facilitate population genomic studies in Ammopiptanthus. The potential drought stress related transcripts identified in this study provide a good start for further investigation into the drought adaptation in Ammopiptanthus.
Histogram presentation of Gene Ontology classification
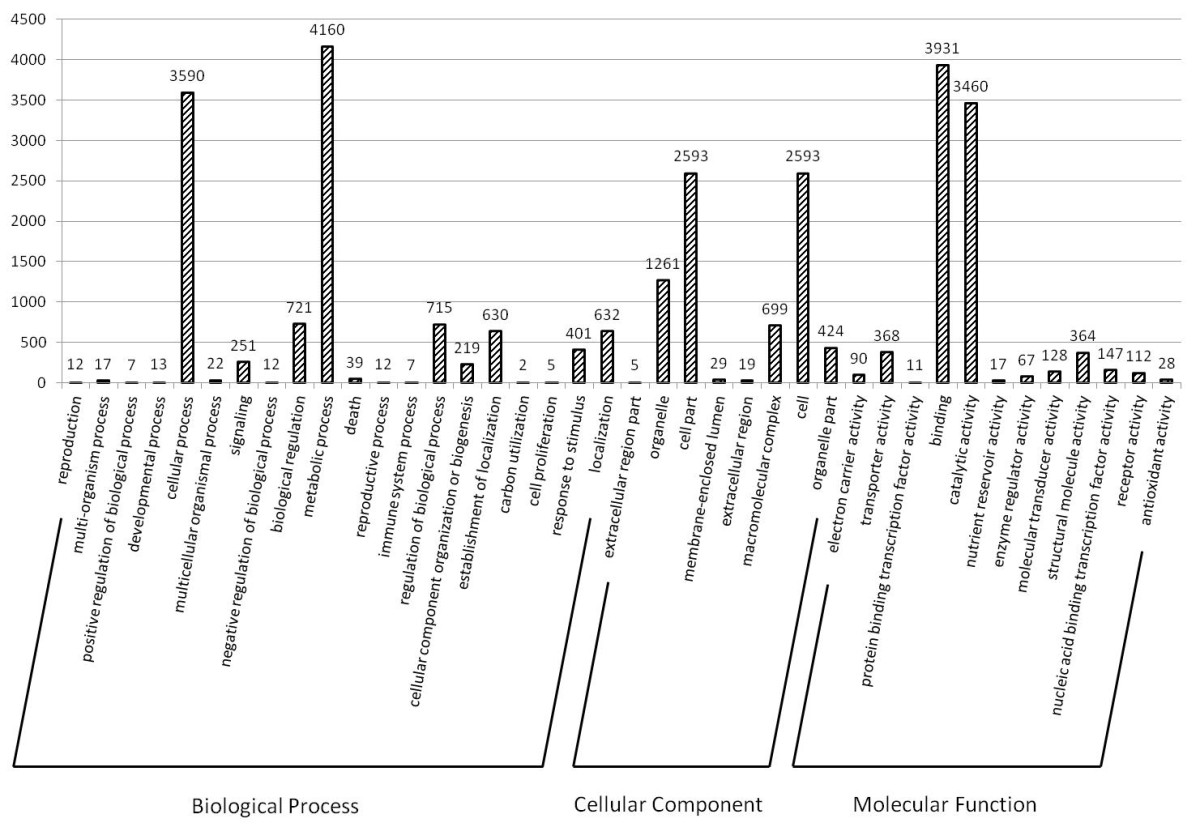 The results are summarized in three main categories: biological process, cellular component, and molecular function. The right y-axis indicates the number of genes in a category. The left y-axis indicates number of unique sequences in a specific category.
The results are summarized in three main categories: biological process, cellular component, and molecular function. The right y-axis indicates the number of genes in a category. The left y-axis indicates number of unique sequences in a specific category.
Related Service
Total RNA Sequencing Service – Total RNA-Seq (Whole Transcriptome) sequencing captures a broader range of gene expression changes and enables the detection of novel transcripts in both coding and non-coding RNA species. Ribo-Zero ribosomal RNA reduction chemistry is used in place a poly-A tail selection which efficiently removes ribosomal RNA (rRNA) This process minimizes ribosomal contamination and optimizes the percentage of reads covering RNA species of interest.…[Learn more…]
References
Zhou Y, Gao F, Liu R, Feng J, Li H, (2012) De novo sequencing and analysis of root transcriptome using 454 pyrosequencing to discover putative genes associated with drought tolerance in Ammopiptanthus mongolicus. BMC Genomics 13(1), 266. [article]
