MicroRNAs (miRNAs) play important regulatory roles in plant development and stress responses. Tomato is an economically important vegetable crop in the world with publicly available genomic information database, but only a limited number of tomato miRNAs have been identified. In a recent study, headed by researchers from Zhejiang Gongshang University, two independent small RNA libraries from mock and Cucumber mosaic virus (CMV)-infected tomatoes were constructed, respectively, and sequenced with using LC Sciences’ miRNA sequencing service. Based on sequence analysis and hairpin structure prediction, a total of 50 plant miRNAs and 273 potentially candidate miRNAs (PC-miRNAs) were firstly identified in tomato, with 12 plant miRNAs and 82 PC-miRNAs supported by both the 3p and 5p strands. Comparative analysis revealed that 79 miRNAs (including 15 new tomato miRNAs) and 40 PC-miRNAs were differentially expressed between the two libraries, and the expression patterns of some new tomato miRNAs and PC-miRNAs were further validated by qRT-PCR. Moreover, potential targets for some of the known and new tomato miRNAs were identified by the recently developed degradome sequencing approach, and target annotation indicated that they were involved in multiple biological processes, including transcriptional regulation and virus resistance. Gene ontology analysis of these target transcripts demonstrated that defense response- and photosynthesis-related genes were most affected in CMV-Fny-infected tomatoes. Because tomato is not only an important crop but also a genetic model for basic biology research, this study contributes to the understanding of miRNAs in response to virus infection.
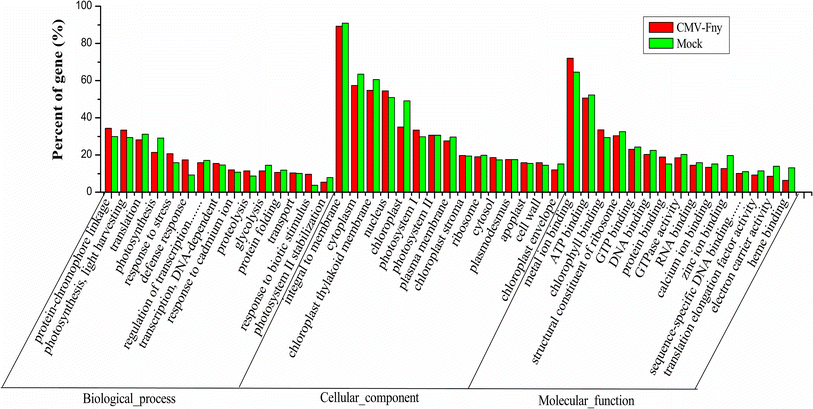
The distribution of top 15 enriched GO terms in biology process, cellular component and molecular function in mock and CMV-Fny-infected libraries. Red bars indicate the enrichment of the GO terms in CMV-Fny-infected library, and green bars indicate the enrichment of the GO terms in mock
Related Service
miRNA/Small RNA Sequencing Service – miRNA sequencing is a new method and powerful tool to identify and quantitatively decode the entire population of microRNAs in your sample. LC Sciences now provides a comprehensive microRNA sequencing service. [Learn more…]
References
Caroline Junli Feng, Shasha Liu, Mengna Wang, Qiulei Lang, Chunzhi Jin Identification of microRNAs and their targets in tomato infected with Cucumber mosaic virus based on deep sequencing (2014) Planta doi: 10.1007/s00425-014-2158-3 [article]
