Aflatoxin contamination is a major constraint in food production worldwide. In peanut (Arachis hypogaea L.), these toxic and carcinogenic aflatoxins are mainly produced by Aspergillus flavus Link and A. parasiticus Speare. The use of RNA interference (RNAi) is a promising method to reduce or prevent the accumulation of aflatoxin in peanut seed.
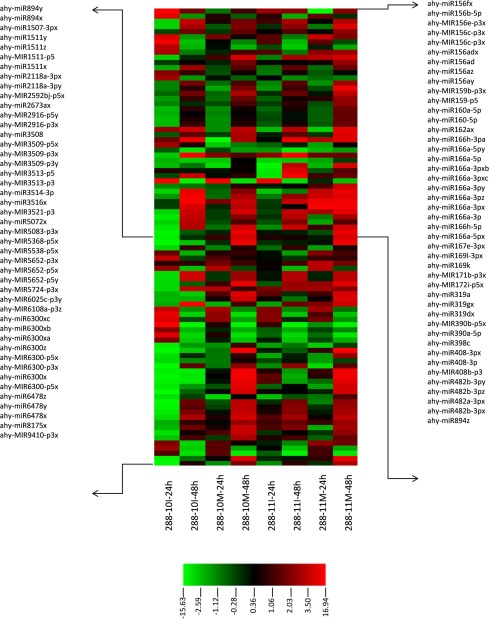
In a recent study published by the National Peanut Research Laboratory, researchers performed high-throughput sequencing of small RNA populations in a control line and in two transformed peanut lines that expressed an inverted repeat targeting five genes involved in the aflatoxin-biosynthesis pathway and that showed up to 100% less aflatoxin B1 than the controls.
The objective of their study was to determine the putative involvement of the small RNA populations in aflatoxin reduction. In total, 41 known microRNA (miRNA) families and many novel miRNAs were identified. Among those, 89 known and 10 novel miRNAs were differentially expressed in the transformed lines. They furthermore found two small interfering RNAs derived from the inverted repeat, and 39 sRNAs that mapped without mismatches to the genome of A. flavus and were present only in the transformed lines.
This information increases our understanding of the effectiveness of RNAi and enables the possible improvement of the RNAi technology for the control of aflatoxins.
Related Service
miRNA/Small RNA Sequencing Service – miRNA sequencing is a new method and powerful tool to identify and quantitatively decode the entire population of microRNAs in your sample. LC Sciences now provides a comprehensive microRNA sequencing service. [Learn more…]
References
I. L. Power, P. M. Dang, V. S. Sobolev, V. A. Orner, J. L. Powell, M. C. Lamb, R. S. Arias (2017) Characterization of small RNA populations in non-transgenic and aflatoxin-reducing-transformed peanut Plant Sci. doi: 10.1016/j.plantsci.2016.12.013 [abstract]
