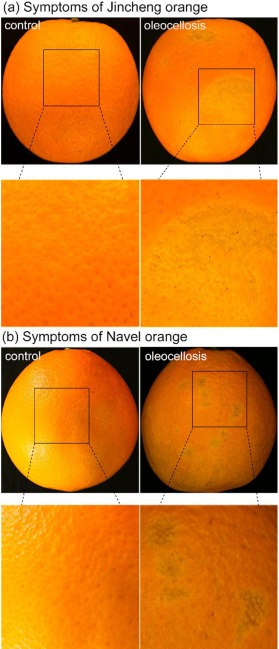
Oleocellosis is a serious physiological disorder in citrus fruit that mainly results in appearance and quality deterioration. It has been well established that the occurrence of oleocellosis is highly correlated with the release of peel oil from citrus fruit, but there is little information on the dynamic changes in the content of volatile constituents and expression of genes involved in terpenoid metabolism during oleocellosis development.
In recent research by scientists from Southwest University, China large changes in the volatile profiles and gene expression in terpenoid metabolism were observed in oleocellosis peels compared to healthy ones. Among volatiles, the decreased contents of α-pinene, d-limonene, β-myrcene, linalool, β-caryophyllene, α-terpineol, nonanal, neryl acetate and (−)-carvone played a major role in these changes. For gene expressions in terpenoid metabolism, the up-regulated genes aldehyde dehydrogenase (NAD+) (ALDH) and the down-regulated genes β-caryophyllene synthase 1 (BCS1), α-terpineol synthase 2 (TES2) and myrcene synthase (MS) were the main differences in oleocellosis peels.
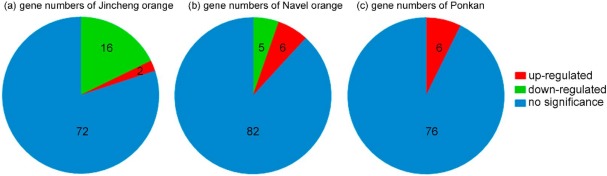
Overview of the expression pattern of the genes involved in terpenoid metabolism of citrus fruit peel: Jincheng orange (a), Navel orange (b), and Ponkan mandarin (c). Genes were considered to be significant with log 2-fold change >1.00 and p < .05.
Related Service
RNA Sequencing Service – LC Sciences provides a one-stop solution (i.e. from sample to data) for RNA-Seq using the latest in rna sequencing technology. The RNA-Seq services provided by LC Sciences are comprehensive and provide the most complete picture of RNA content in your samples. We can help you set up and conduct a high-quality, well-controlled RNA-Seq experiment based on the latest deep-sequencing technologies [Learn more…]
References
J. Xie, L. Deng, Y. Zhou, S. Yao, K. Zeng (2017) Analysis of changes in volatile constituents and expression of genes involved in terpenoid metabolism in oleocellosis peel Food Chem. doi: 10.1016/j.foodchem.2017.09.142 [abstract]